Robust Counterfactual Medical Image Generation
Counterfactual images represent what an image would have looked like if it had different specified characteristics. They find use in a wide range of tasks in medical imaging:
| Counterfactual variable | Task |
|---|---|
| Age / time | Forecasting |
| Phenotype | Data exploration |
| Outcome | Biomarker visualization |
| Feature | Feature visualization |
| Acquisition settings | Modality transfer |
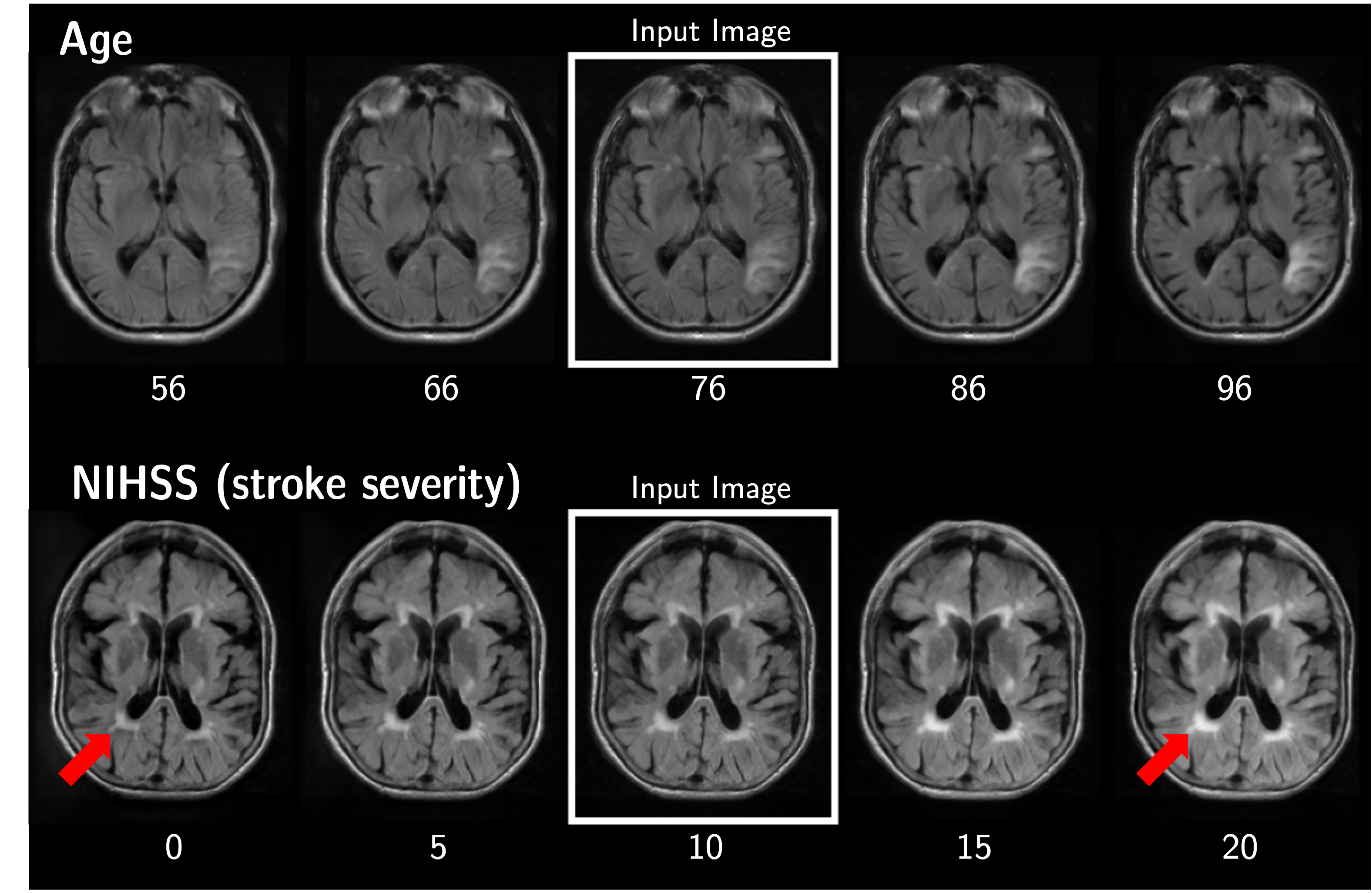
Generative models offer a powerful tool for generating counterfactual images. But we want such models to make the minimal changes necessary to the input image, whereas existing models usually change aspects of the input image unrelated to the counterfactual variable.

These problems are exacerbated in small, real-world (clinical) datasets, where a generative model is more susceptible to artifacts and differences in scanner settings.
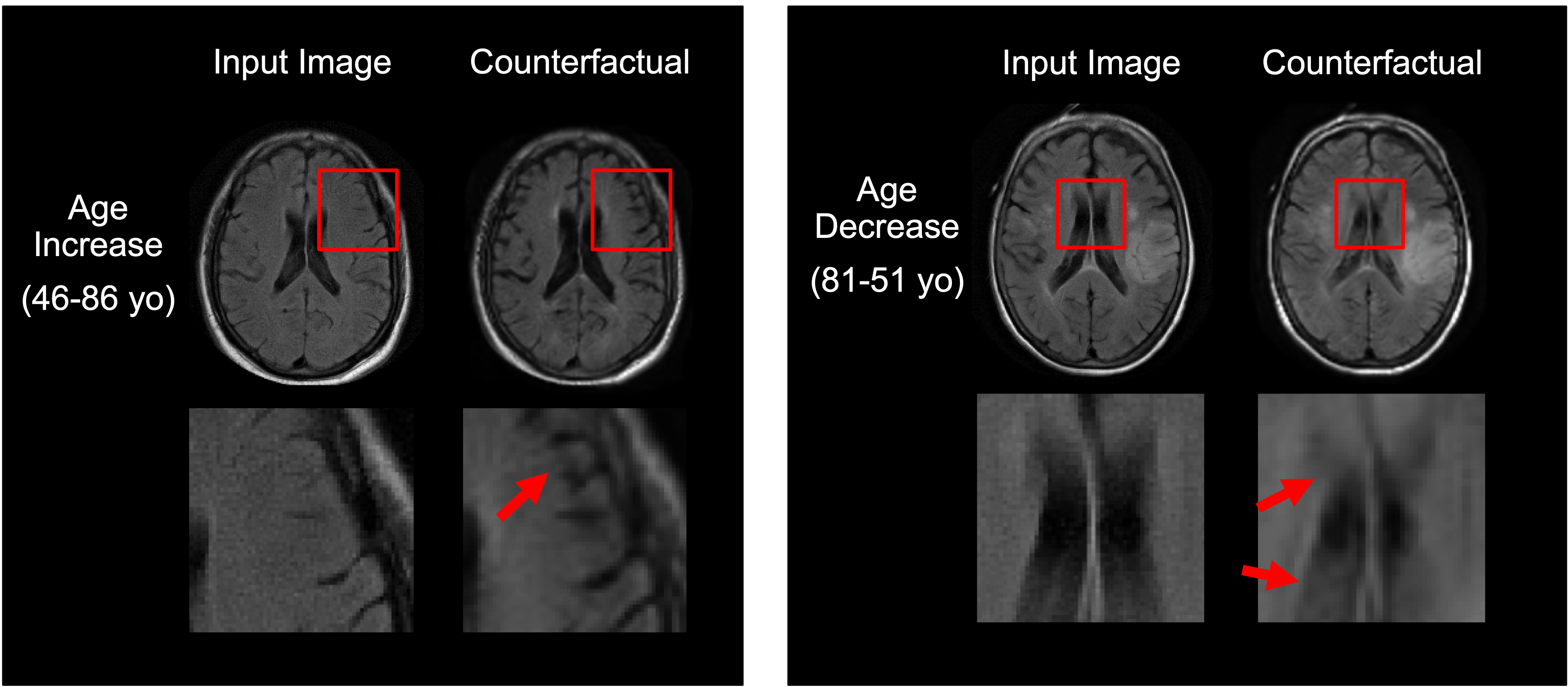
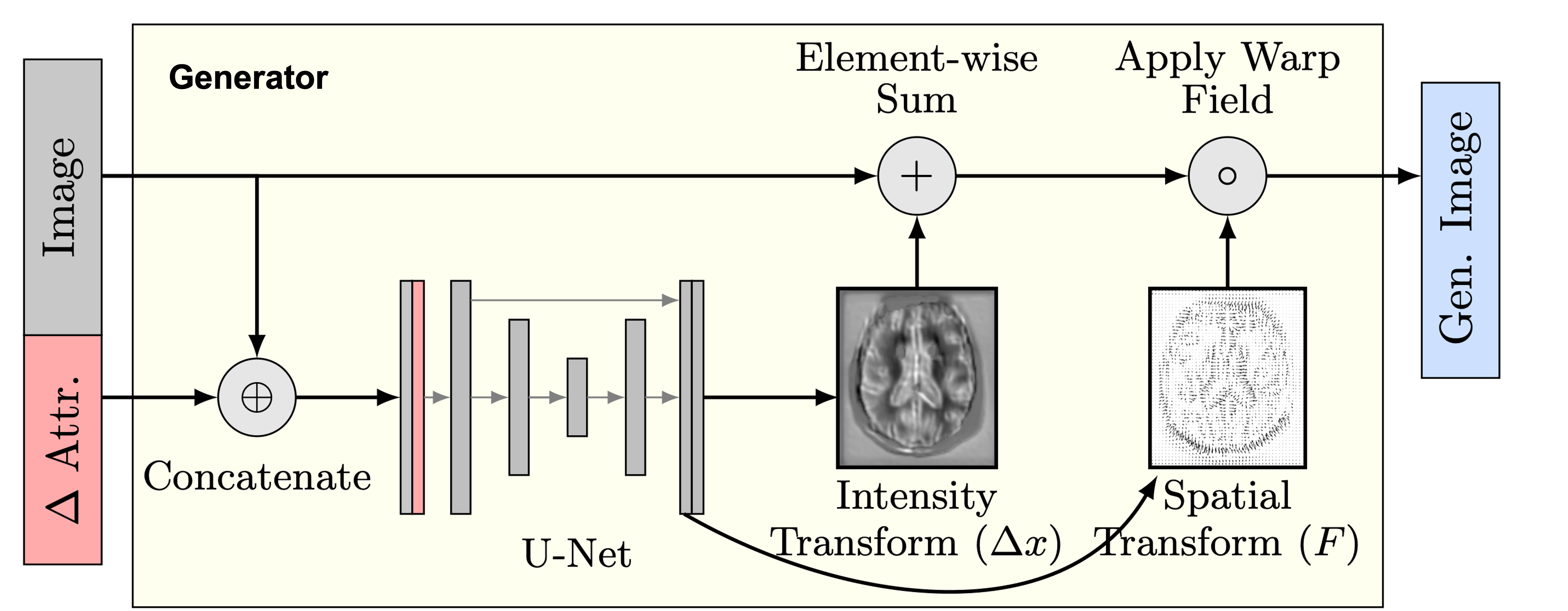
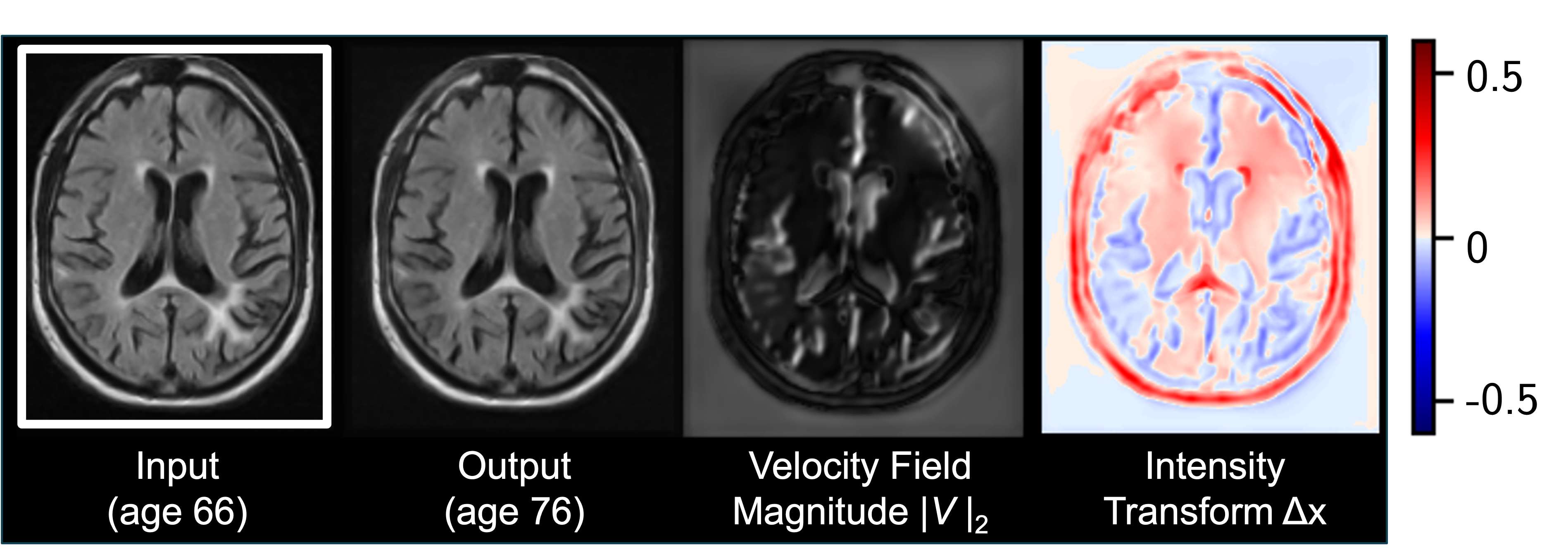
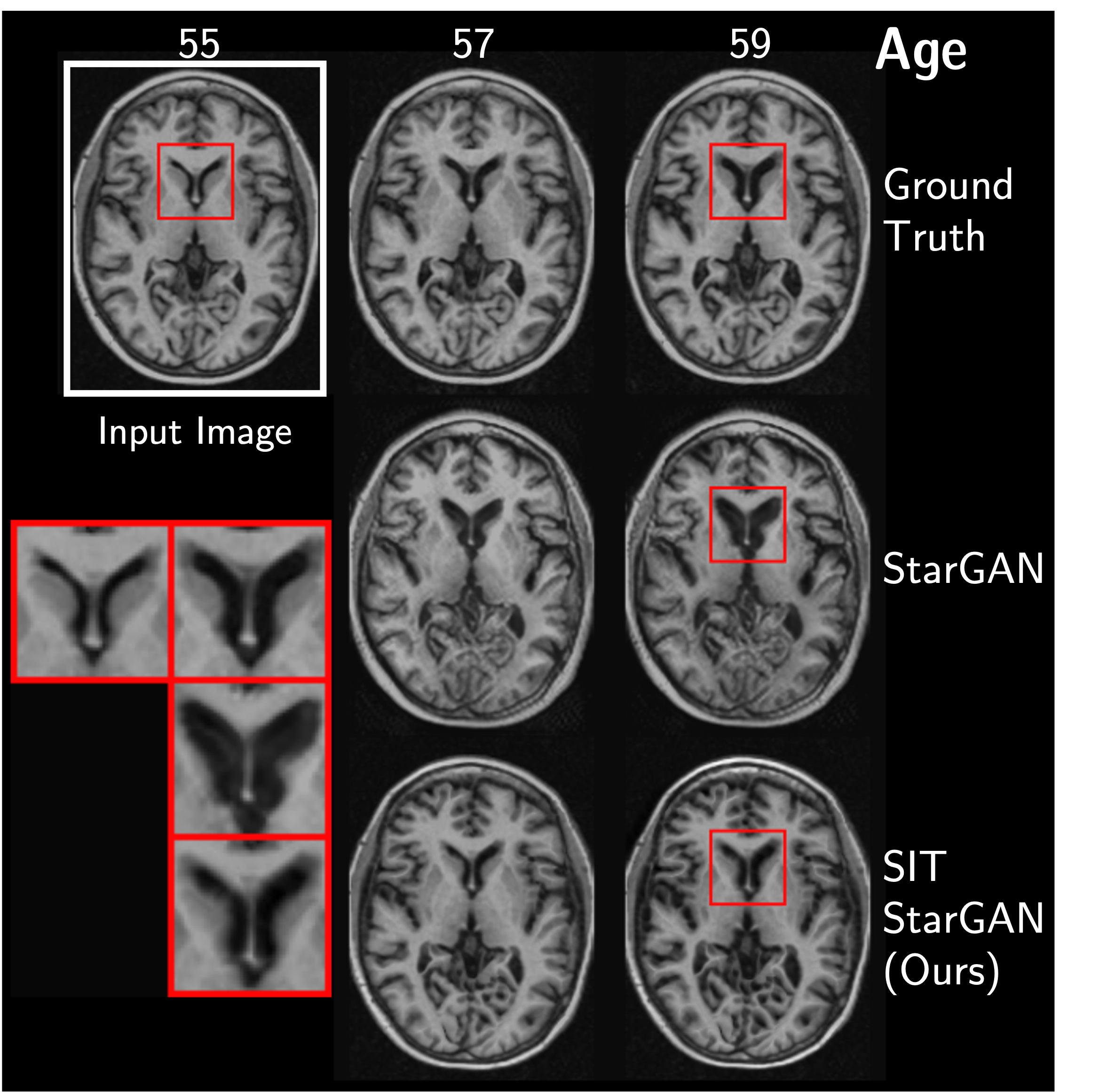
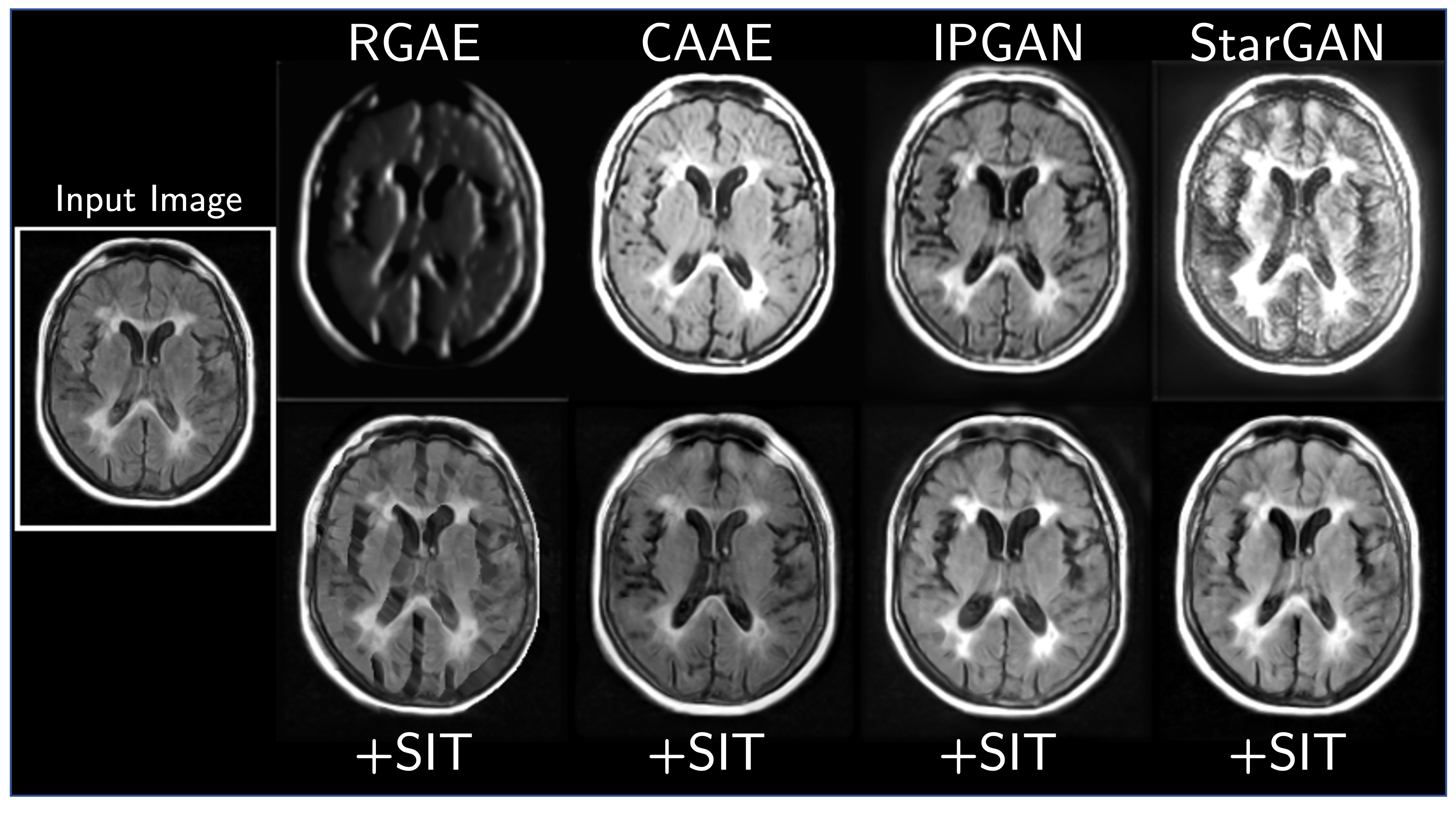
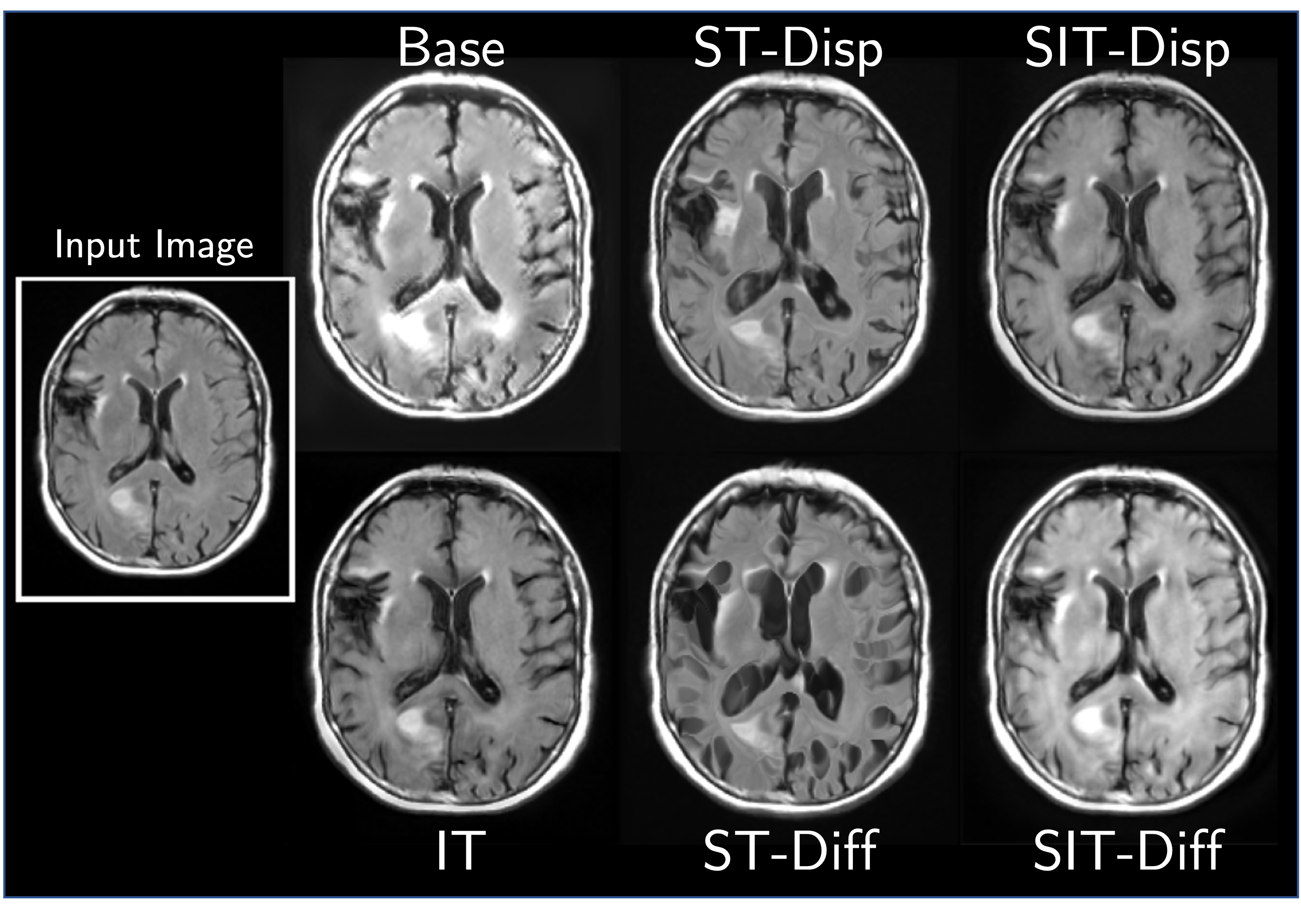
We address this problem by introducing spatial-intensity transforms. We parameterize the output of the generator in terms of a smooth deformation field and sparse intensity transform. The smoothness and sparsity can be controlled flexibly for different applications.
We find that this simple construction confers improved robustness and image quality on a variety of architectures, counterfactual variables, and datasets. It also disentangles morphological changes from tissue intensity and textural changes.
Read our MICCAI paper for more details, and try out our code.
More Results
Bibtex
@inproceedings{wang2020spatial,
title={Spatial-intensity transform GANs for high fidelity medical image-to-image translation},
author={Wang, Clinton J and Rost, Natalia S and Golland, Polina},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={749--759},
year={2020},
organization={Springer}
}